Research
Research in the Panda lab is focused in these areas:
Sleep
Sleep, alongside exercise and diet, stands as a foundational pillar of health. The intricate dance of circadian behaviors plays a crucial role in determining the quality of our sleep, which in turn, significantly impacts our physiological performance. At the Panda Lab, we are deeply fascinated by the dynamic interplay between these elements. We delve into questions such as the influence of sleep on immune response and the effects of dietary timing on sleep quality.
Utilizing a blend of sophisticated tools, our laboratory is at the forefront of sleep research. We employ electroencephalograms (EEG) and electromyograms (EMG) to meticulously assess sleep patterns in mice, offering us a window into the neural underpinnings of sleep. Additionally, we use activity monitoring to deduce the sleep/wake cycles in drosophila, providing a broader perspective on sleep behaviors across species. Our behavioral mouse models are instrumental in rigorously examining how sleep impacts various physiological aspects, from metabolic processes to immune functions.
Our approach is comprehensive, integrating physiological measurements and perturbations with multi-omics techniques. This allows us to unravel and synthesize how sleep influences organ physiology and molecular interactions. By shedding light on the molecular mechanisms underpinning sleep, we aim to pave the way for future therapeutics. This mission is more critical than ever in a society where the importance of sleep is often overlooked, yet its implications on health are profound. Through our research at the Panda Lab, we strive to elevate the understanding and appreciation of sleep’s pivotal role in our well-being.
Exercise
Physical performance is a critical factor in promoting both longevity and healthspan, requiring contributions from nearly every system in the body (Figure 1). Exercise—whether it be resistance training, endurance exercise, or high-intensity interval training—provides far-reaching physiological benefits, impacting areas such as metabolism, insulin sensitivity, neurodegenerative disease prevention, and cancer progression. By unraveling the molecular mechanisms behind optimal physical performance, we aim to uncover discoveries that can significantly benefit public health.
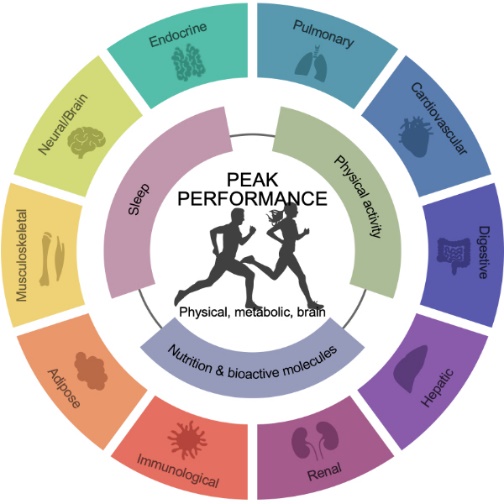
Figure 1. Factors influencing performance.
In the lab of Satchin Panda, we explore key questions at the intersection of exercise and molecular biology. Our research investigates the effects of voluntary wheel running on both multi-organ and single-cell levels. Through this work, we have identified key physiological pathways that are differentially expressed in response to exercise in both male and female mice (Figure 2). Additionally, we are studying how sleep disruption affects metabolism and physical performance, and how time-restricted feeding (TRF) with varying macronutrient profiles can influence exercise capacity in male and female preclinical settings models.
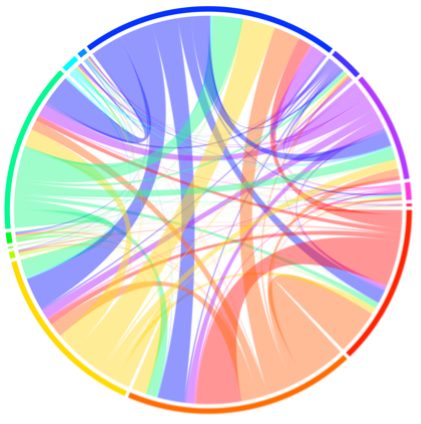
Figure 2. Multi-tissue transcriptomic response to exercise.
To answer these questions, we employ a comprehensive approach that integrates treadmill performance, force generation, behavioral assessments, physiological measurements, and biochemical analyses. Our lab also harnesses cutting-edge -omics techniques—including spatial transcriptomics, bulk RNA sequencing, single-cell gene expression, and metabolomics—to explore the molecular benefits of exercise. A distinguishing feature of our research is our ability to analyze and integrate data from multiple organ systems in collaboration with an extensive network of expert partners.
Metabolism
Circadian rhythms play a critical role in regulating various physiological functions, including metabolism. Metabolic processes, such as energy expenditure, nutrient digestion, absorption and utilization, and hormone production, are often synchronized with circadian rhythms. Disruptions to circadian rhythms, caused by factors such as mistimed eating, shift work, jet lag, or exposure to excessive light at night, can increase the risk of developing metabolic disorders. Investigating mechanisms that can maintain and bolster these circadian rhythms in metabolism is important for improving health and preventing age-associated diseases such as obesity, type-2 diabetes, neurodegenerative diseases, cardiovascular disease, and cancer.
In the lab, we are investigating the interplay between the timing of nutrition and circadian rhythms. Time-restricted feeding (TRF) is a feeding pattern intervention in which organisms are fed for 6-12 hours during their active phase in a daily, consistent manner. The current lab projects involve various fly and rodent models of diseases such as obesity, type-2 diabetes, cardiovascular disease, fatty liver disease, cancer, and aging to understand the role of daily feeding-fasting cycles and bolstering circadian rhythms via TRF in improving health. We are also studying the role of sex differences and aging in regulating circadian rhythms in physiology and metabolism, and how it affects TRF response.
To address these questions, we use several behavioral, physiological, pathological, and biochemical tests and measurements. We also use state-of-the-art omics technologies such as genomics, epigenomics, single-cell/nuclei and spatial gene expression, and lipidomics/metabolomics to understand the mechanisms of TRF. Moreover, since metabolism involves changes in nutrient flux and endocrine regulation between different tissues/organs, we investigate these molecular mechanisms across multiple organ systems.
Neuroscience
Our laboratory studies the neuroscience aspects of the regulation by light of circadian rhythms in mammals, in health and disease.
These projects are conducted on multiple scales:
- Using a combination of fluorescent and electron microscopy imaging, we characterize the morphology of retina cells and brain regions receiving direct input from the retina. We are particularly interested in the non-visual opsins and the suprachiasmatic nucleus (SCN).
- We use state-of-the-art electron microscopy to elucidate the connectomics of our regions of interest. We established the retinal network involving the melanopsin-expressing cells, characterized the local specialization of their axon terminal, and revealed the subregion specificity and integration of retinal input of the SCN.
- Finally, we correlate our morphological and connectomics findings with functional assays, in and ex vivo. These assays include evaluating the electrical response to light of retinal photoreceptor cells, the effect of lighting conditions on locomotor activity, and sleep regulation.
Translational Research
The clinical arm of the lab studies how lifestyle interventions (alone or in combination with medication) can improve age-related diseases from pre-diabetes, diabetes, cardiovascular disease, or cancer in humans. We use a combination of cutting-edge technology including wearable devices (ex. continuous glucose monitors and actigraphy) and a smartphone app our lab created (myCircadianClock) to implement and monitor clinical interventions in participants living in the real world. Our research is focused on using personalized circadian interventions, such as time-restricted eating, to help treat diseases such as prediabetes/type 2 diabetes, metabolic syndrome, and affective disorders as well as to help offset the burden of shift work.
Time-Restricted Eating (TRE) is a dietary regimen with a consistent fasting window of at least 14 hours per day in humans and no explicit limit on energy intake during eating hours.
Key Findings from TRE Research
- TRE improves multiple components of metabolic syndrome, including hemoglobin A1c (gold standard for assessing glucose over ~3 months), LDL-cholesterol, and weight. These improvements have been found in many clinical trials including adults with metabolic syndrome and firefighters working 24-hours shifts
- TRE is safe. No serious adverse effects have been resported with 8-10 hour time-restricted eating in adults. It has not been adequately tested in minors or women who are pregnant or breastfeeding.
- TRE can improve self-reported energy and restfulness following sleep.
- TRE studies in humans are based on many studies in mice and flies have have demonstrated widespread health benefits of time-restricted feeding (TRF – feeding in animals models).
Clinical Trials
Ongoing Studies
- Understanding eating patterns world wide. If you 18 or older and have a smartphone, you are eligible to participate in our largest research trial, no matter where you live. All you have to do is download the myCircadianClock app on your app store and start logging when you eat. For for the first 2 weeks, we want to know your currently lifestyle. After that, you can set goals for when you eat and sleep to transform your health. You can even log you health factors so we can better understand how eating patterns impact health on a global scale. For more information, go to mycircadianclock.org.
- Time-Restricted Eating for Type 2 Diabetes (TRE-T2D). This study aims to assess the health benefits TRE in adutls with Type 2 Diabetes. Participation is limited to adults in the San Diego Area. If you are interested in participating, you can contact our study coordinators at (858) 246-2406 or preventivecvresearch@health.ucsd.edu. This study is funded by the Larry L. Hillblom Foundation.
Coming Soon:
- The SHIFT Study. This sudy aims to optimize the timing of eating to help reduce the risk of diseases in shift workers. Particpation in open to nurses and nursing assistants to do night shift work in the San Diego area. If you are interested in participating, contact us at: research@mycircadianclock.org or call us as 858-453-4100 x 1077. This study is funded by The National Institute for Diabetes and Disease of the Kidney (NIDDK) of the National Institute of Health (NIH).
Data Science
Our lab, along with valued collaborators, generates vast multi-omics datasets spanning DNA-Seq, RNA-Seq, proteomics, metabolomics, phenotypes, and clinical trial data. To manage this wealth of information, we have developed scalable data processing pipelines that ensure uniform analysis across high-performance computing clusters and cloud-based systems. We integrate these multi-omics datasets using advanced statistical models to answer critical research questions. Committed to open data principles, we also create user-friendly data accessioning and visualization web applications to enhance accessibility.
Our lab generates vast multi-omics datasets, including DNA-Seq, RNA-Seq, proteomics, metabolomics, phenotypes, and clinical trials. To handle this data, we have developed scalable pipelines for uniform analysis across high-performance clusters and cloud systems, integrating advanced statistical models to answer critical research questions. We also prioritize open data principles, providing user-friendly data accession and visualization tools.
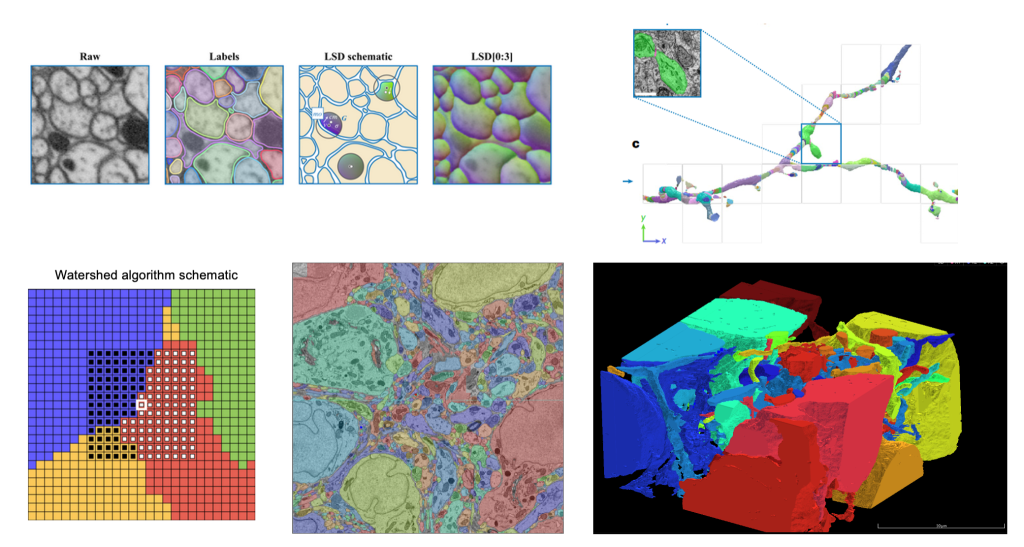
We have enhanced our computational toolkit by deploying computer vision models such as Local Shape Descriptors to analyze large-scale brain imaging data, significantly improving segmentation accuracy and efficiency.
Our work also extends into using Multi-modal Large Language Models for visual classification of food log images, validating user entries with high accuracy. We are also building an experimental AI chatbot to act as an expert in Circadian Rhythm research. By leveraging modern techniques like Retrieval Augmented Generation using a vector database of scientific literature, we can improve information retrieval, research productivity, and scientific outreach.
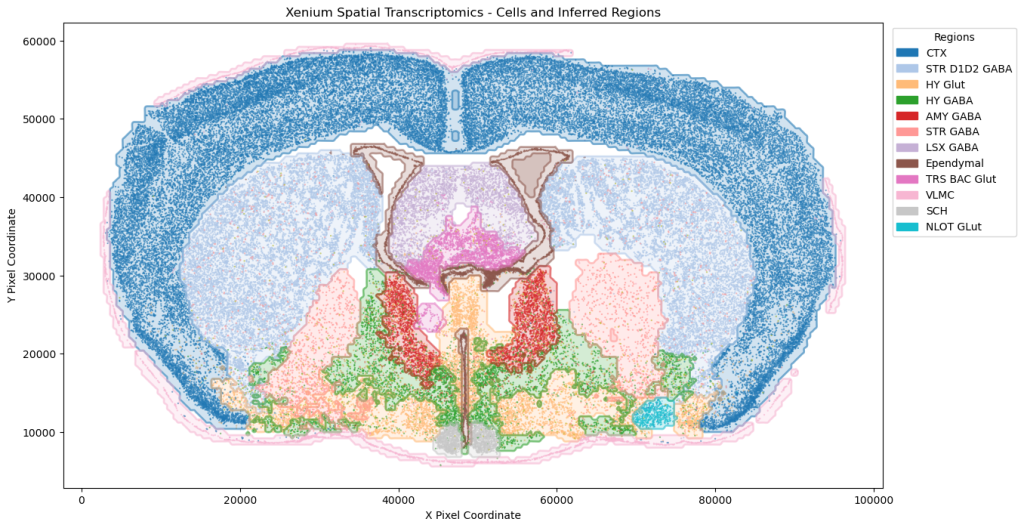
On the transcriptomics front, our single-cell RNA analysis on sleep disruption studies, supported by PySCENIC, has revealed transcription factors linked to immune response variations. We have expanded into the paradigm Xenium spatial transcriptomics, utilizing advanced computer vision techniques for cell type annotation and insights into brain architecture.
In an effort to enable widespread data accessibility we’ve developed a data visualization web application for comparing cyclical gene expression patterns, aiding in understanding circadian regulation across experiments.
Aging, Cancer and Alzheimer’s Disease
Age is the major risk factor for many diseases, including cancer and neurodegeneration. Alzheimer’s disease (AD) is a progressive neurodegenerative disease. It usually starts with mild memory loss, through cognition difficulty, and ends up with a complete loss of perception and response to environmental stimuli. In 2023, there were 6.7 million Americans age 65 or older living with AD. To better our understanding of AD pathology, its impact on circadian rhythm, the underlying molecular basis, and also to explore the potential benefits of Time-restricted eating on AD, we are currently conducting experiments in different animal models of AD. We utilize a variety of research tools including genetic, behavioral, molecular, immunohistological, electrophysiological, and biochemical to investigate AD progression in animals. Our ultimate goal is to find ways to slow down AD progression and improve the quality of life in AD patients. Circadian disruption is linked to an increased risk of cancer. Cell and animal models have been established to elucidate the intricate molecular interactions between circadian rhythm regulation and the development of cancer, as well as the potential implications for cancer therapy.
Interestingly, our current results show that time-restricted feeding (TRF) in animal models of AD is able to significantly improve the mobility and extend the lifespan of the treated animals. Additionally, we are testing the potential function of TRF in cognition improvement and memory preservation. With the assistance from next-generation sequencing technology and sophisticated genetic analysis, we are trying to pin down the underlying genes for TRF benefits in AD animal models. Those results will improve our knowledge base about AD pathology, illustrate the molecular mechanism involved in TRF benefits, and hopefully provide promising pharmaceutical targeting sites to slow down the progress of AD.